Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Template-based prediction.¶
In this tutorial, we show how to improve inter-subject similarity using a template computed across multiple source subjects. For this purpose, we create a template using Procrustes alignment (hyperalignment) to which we align the target subject, using shared information. We then compare the voxelwise similarity between the target subject and the template to the similarity between the target subject and the anatomical Euclidean average of the source subjects.
We mostly rely on Python common packages and on nilearn to handle functional data in a clean fashion.
To run this example, you must launch IPython via ipython
--matplotlib in a terminal, or use jupyter-notebook.
Retrieve the data¶
In this example we use the IBC dataset, which includes a large number of different contrasts maps for 12 subjects. We download the images for subjects sub-01, sub-02, sub-04, sub-05, sub-06 and sub-07 (or retrieve them if they were already downloaded). imgs is the list of paths to available statistical images for each subjects. df is a dataframe with metadata about each of them. mask is a binary image used to extract grey matter regions.
[_add_readme_to_default_data_locations] Added README.md to
/home/runner/nilearn_data
[get_dataset_dir] Dataset created in /home/runner/nilearn_data/ibc
[fetch_single_file] Downloading data from https://osf.io/pcvje/download ...
[fetch_single_file] ...done. (1 seconds, 0 min)
[uncompress_file] Extracting data from
/home/runner/nilearn_data/ibc/8e275a34345802c5c273312d85957d6c/download...
[uncompress_file] .. done.
[fetch_single_file] Downloading data from https://osf.io/yvju3/download ...
[fetch_single_file] ...done. (3 seconds, 0 min)
[uncompress_file] Extracting data from
/home/runner/nilearn_data/ibc/fe06df963fcb3fd454f63a33f0864e8d/download...
[uncompress_file] .. done.
[fetch_single_file] Downloading data from https://osf.io/8z23h/download ...
[_chunk_report_] Downloaded 802816 of 21185337 bytes (3.8%%, 27.0s remaining)
[_chunk_report_] Downloaded 1613824 of 21185337 bytes (7.6%%, 25.6s remaining)
[_chunk_report_] Downloaded 2416640 of 21185337 bytes (11.4%%, 24.5s
remaining)
[_chunk_report_] Downloaded 3227648 of 21185337 bytes (15.2%%, 23.4s
remaining)
[_chunk_report_] Downloaded 4038656 of 21185337 bytes (19.1%%, 22.3s
remaining)
[_chunk_report_] Downloaded 4849664 of 21185337 bytes (22.9%%, 21.2s
remaining)
[_chunk_report_] Downloaded 5652480 of 21185337 bytes (26.7%%, 20.2s
remaining)
[_chunk_report_] Downloaded 6463488 of 21185337 bytes (30.5%%, 19.1s
remaining)
[_chunk_report_] Downloaded 7274496 of 21185337 bytes (34.3%%, 18.1s
remaining)
[_chunk_report_] Downloaded 8077312 of 21185337 bytes (38.1%%, 17.0s
remaining)
[_chunk_report_] Downloaded 8888320 of 21185337 bytes (42.0%%, 16.0s
remaining)
[_chunk_report_] Downloaded 9699328 of 21185337 bytes (45.8%%, 14.9s
remaining)
[_chunk_report_] Downloaded 10510336 of 21185337 bytes (49.6%%, 13.9s
remaining)
[_chunk_report_] Downloaded 11321344 of 21185337 bytes (53.4%%, 12.8s
remaining)
[_chunk_report_] Downloaded 12124160 of 21185337 bytes (57.2%%, 11.8s
remaining)
[_chunk_report_] Downloaded 12926976 of 21185337 bytes (61.0%%, 10.7s
remaining)
[_chunk_report_] Downloaded 13737984 of 21185337 bytes (64.8%%, 9.7s
remaining)
[_chunk_report_] Downloaded 14548992 of 21185337 bytes (68.7%%, 8.6s
remaining)
[_chunk_report_] Downloaded 15351808 of 21185337 bytes (72.5%%, 7.6s
remaining)
[_chunk_report_] Downloaded 16162816 of 21185337 bytes (76.3%%, 6.5s
remaining)
[_chunk_report_] Downloaded 16973824 of 21185337 bytes (80.1%%, 5.5s
remaining)
[_chunk_report_] Downloaded 17784832 of 21185337 bytes (83.9%%, 4.4s
remaining)
[_chunk_report_] Downloaded 18595840 of 21185337 bytes (87.8%%, 3.4s
remaining)
[_chunk_report_] Downloaded 19406848 of 21185337 bytes (91.6%%, 2.3s
remaining)
[_chunk_report_] Downloaded 20217856 of 21185337 bytes (95.4%%, 1.3s
remaining)
[_chunk_report_] Downloaded 21020672 of 21185337 bytes (99.2%%, 0.2s
remaining)
[fetch_single_file] ...done. (29 seconds, 0 min)
[uncompress_file] Extracting data from
/home/runner/nilearn_data/ibc/100352739b7501f0ed04920933b4be36/download...
[uncompress_file] .. done.
[fetch_single_file] Downloading data from https://osf.io/e9kbm/download ...
[_chunk_report_] Downloaded 9207808 of 21196887 bytes (43.4%%, 1.3s
remaining)
[fetch_single_file] ...done. (4 seconds, 0 min)
[uncompress_file] Extracting data from
/home/runner/nilearn_data/ibc/331a0a579c6e46c0911502a96215b358/download...
[uncompress_file] .. done.
[fetch_single_file] Downloading data from https://osf.io/qn5b6/download ...
[_chunk_report_] Downloaded 868352 of 21197218 bytes (4.1%%, 23.5s remaining)
[_chunk_report_] Downloaded 1613824 of 21197218 bytes (7.6%%, 24.6s remaining)
[_chunk_report_] Downloaded 2424832 of 21197218 bytes (11.4%%, 23.8s
remaining)
[_chunk_report_] Downloaded 3235840 of 21197218 bytes (15.3%%, 22.9s
remaining)
[_chunk_report_] Downloaded 4046848 of 21197218 bytes (19.1%%, 21.9s
remaining)
[_chunk_report_] Downloaded 4857856 of 21197218 bytes (22.9%%, 20.9s
remaining)
[_chunk_report_] Downloaded 5660672 of 21197218 bytes (26.7%%, 20.0s
remaining)
[_chunk_report_] Downloaded 6471680 of 21197218 bytes (30.5%%, 18.9s
remaining)
[_chunk_report_] Downloaded 7274496 of 21197218 bytes (34.3%%, 17.9s
remaining)
[_chunk_report_] Downloaded 8085504 of 21197218 bytes (38.1%%, 16.9s
remaining)
[_chunk_report_] Downloaded 8896512 of 21197218 bytes (42.0%%, 15.8s
remaining)
[_chunk_report_] Downloaded 9707520 of 21197218 bytes (45.8%%, 14.8s
remaining)
[_chunk_report_] Downloaded 10518528 of 21197218 bytes (49.6%%, 13.8s
remaining)
[_chunk_report_] Downloaded 11329536 of 21197218 bytes (53.4%%, 12.7s
remaining)
[_chunk_report_] Downloaded 12132352 of 21197218 bytes (57.2%%, 11.7s
remaining)
[_chunk_report_] Downloaded 12943360 of 21197218 bytes (61.1%%, 10.7s
remaining)
[_chunk_report_] Downloaded 13754368 of 21197218 bytes (64.9%%, 9.6s
remaining)
[_chunk_report_] Downloaded 14565376 of 21197218 bytes (68.7%%, 8.6s
remaining)
[_chunk_report_] Downloaded 15376384 of 21197218 bytes (72.5%%, 7.5s
remaining)
[_chunk_report_] Downloaded 16187392 of 21197218 bytes (76.4%%, 6.5s
remaining)
[_chunk_report_] Downloaded 16998400 of 21197218 bytes (80.2%%, 5.4s
remaining)
[_chunk_report_] Downloaded 17809408 of 21197218 bytes (84.0%%, 4.4s
remaining)
[_chunk_report_] Downloaded 18620416 of 21197218 bytes (87.8%%, 3.3s
remaining)
[_chunk_report_] Downloaded 19431424 of 21197218 bytes (91.7%%, 2.3s
remaining)
[_chunk_report_] Downloaded 20242432 of 21197218 bytes (95.5%%, 1.2s
remaining)
[_chunk_report_] Downloaded 21045248 of 21197218 bytes (99.3%%, 0.2s
remaining)
[fetch_single_file] ...done. (29 seconds, 0 min)
[uncompress_file] Extracting data from
/home/runner/nilearn_data/ibc/cd396fed594eb866baecd48b70ddf7e7/download...
[uncompress_file] .. done.
[fetch_single_file] Downloading data from https://osf.io/u74a3/download ...
[_chunk_report_] Downloaded 8216576 of 21185350 bytes (38.8%%, 1.6s
remaining)
[fetch_single_file] ...done. (3 seconds, 0 min)
[uncompress_file] Extracting data from
/home/runner/nilearn_data/ibc/fc5556cc3678df4f4ab566414382180a/download...
[uncompress_file] .. done.
[fetch_single_file] Downloading data from https://osf.io/83bje/download ...
[_chunk_report_] Downloaded 860160 of 21188335 bytes (4.1%%, 23.6s remaining)
[_chunk_report_] Downloaded 1622016 of 21188335 bytes (7.7%%, 24.5s remaining)
[_chunk_report_] Downloaded 2433024 of 21188335 bytes (11.5%%, 23.7s
remaining)
[_chunk_report_] Downloaded 3235840 of 21188335 bytes (15.3%%, 22.9s
remaining)
[_chunk_report_] Downloaded 4046848 of 21188335 bytes (19.1%%, 21.9s
remaining)
[_chunk_report_] Downloaded 4857856 of 21188335 bytes (22.9%%, 20.9s
remaining)
[_chunk_report_] Downloaded 5668864 of 21188335 bytes (26.8%%, 19.9s
remaining)
[_chunk_report_] Downloaded 6471680 of 21188335 bytes (30.5%%, 18.9s
remaining)
[_chunk_report_] Downloaded 7282688 of 21188335 bytes (34.4%%, 17.9s
remaining)
[_chunk_report_] Downloaded 8093696 of 21188335 bytes (38.2%%, 16.8s
remaining)
[_chunk_report_] Downloaded 8896512 of 21188335 bytes (42.0%%, 15.8s
remaining)
[_chunk_report_] Downloaded 9707520 of 21188335 bytes (45.8%%, 14.8s
remaining)
[_chunk_report_] Downloaded 10518528 of 21188335 bytes (49.6%%, 13.7s
remaining)
[_chunk_report_] Downloaded 11329536 of 21188335 bytes (53.5%%, 12.7s
remaining)
[_chunk_report_] Downloaded 12140544 of 21188335 bytes (57.3%%, 11.6s
remaining)
[_chunk_report_] Downloaded 12943360 of 21188335 bytes (61.1%%, 10.6s
remaining)
[_chunk_report_] Downloaded 13754368 of 21188335 bytes (64.9%%, 9.6s
remaining)
[_chunk_report_] Downloaded 14565376 of 21188335 bytes (68.7%%, 8.5s
remaining)
[_chunk_report_] Downloaded 15376384 of 21188335 bytes (72.6%%, 7.5s
remaining)
[_chunk_report_] Downloaded 16179200 of 21188335 bytes (76.4%%, 6.5s
remaining)
[_chunk_report_] Downloaded 16990208 of 21188335 bytes (80.2%%, 5.4s
remaining)
[_chunk_report_] Downloaded 17801216 of 21188335 bytes (84.0%%, 4.4s
remaining)
[_chunk_report_] Downloaded 18612224 of 21188335 bytes (87.8%%, 3.3s
remaining)
[_chunk_report_] Downloaded 19415040 of 21188335 bytes (91.6%%, 2.3s
remaining)
[_chunk_report_] Downloaded 20226048 of 21188335 bytes (95.5%%, 1.2s
remaining)
[_chunk_report_] Downloaded 21028864 of 21188335 bytes (99.2%%, 0.2s
remaining)
[fetch_single_file] ...done. (29 seconds, 0 min)
[uncompress_file] Extracting data from
/home/runner/nilearn_data/ibc/1beaa1b5a1734a1afbf1c844e1f7a60e/download...
[uncompress_file] .. done.
[fetch_single_file] Downloading data from https://osf.io/43j69/download ...
[_chunk_report_] Downloaded 876544 of 21187400 bytes (4.1%%, 24.6s remaining)
[_chunk_report_] Downloaded 1687552 of 21187400 bytes (8.0%%, 24.4s remaining)
[_chunk_report_] Downloaded 2490368 of 21187400 bytes (11.8%%, 23.7s
remaining)
[_chunk_report_] Downloaded 3301376 of 21187400 bytes (15.6%%, 22.8s
remaining)
[_chunk_report_] Downloaded 4112384 of 21187400 bytes (19.4%%, 21.8s
remaining)
[_chunk_report_] Downloaded 4923392 of 21187400 bytes (23.2%%, 20.8s
remaining)
[_chunk_report_] Downloaded 5726208 of 21187400 bytes (27.0%%, 19.8s
remaining)
[_chunk_report_] Downloaded 6537216 of 21187400 bytes (30.9%%, 18.8s
remaining)
[_chunk_report_] Downloaded 7348224 of 21187400 bytes (34.7%%, 17.8s
remaining)
[_chunk_report_] Downloaded 8159232 of 21187400 bytes (38.5%%, 16.8s
remaining)
[_chunk_report_] Downloaded 8970240 of 21187400 bytes (42.3%%, 15.7s
remaining)
[_chunk_report_] Downloaded 9781248 of 21187400 bytes (46.2%%, 14.7s
remaining)
[_chunk_report_] Downloaded 10592256 of 21187400 bytes (50.0%%, 13.6s
remaining)
[_chunk_report_] Downloaded 11395072 of 21187400 bytes (53.8%%, 12.6s
remaining)
[_chunk_report_] Downloaded 12206080 of 21187400 bytes (57.6%%, 11.6s
remaining)
[_chunk_report_] Downloaded 13017088 of 21187400 bytes (61.4%%, 10.5s
remaining)
[_chunk_report_] Downloaded 13828096 of 21187400 bytes (65.3%%, 9.5s
remaining)
[_chunk_report_] Downloaded 14630912 of 21187400 bytes (69.1%%, 8.5s
remaining)
[_chunk_report_] Downloaded 15441920 of 21187400 bytes (72.9%%, 7.4s
remaining)
[_chunk_report_] Downloaded 16252928 of 21187400 bytes (76.7%%, 6.4s
remaining)
[_chunk_report_] Downloaded 17063936 of 21187400 bytes (80.5%%, 5.3s
remaining)
[_chunk_report_] Downloaded 17874944 of 21187400 bytes (84.4%%, 4.3s
remaining)
[_chunk_report_] Downloaded 18677760 of 21187400 bytes (88.2%%, 3.2s
remaining)
[_chunk_report_] Downloaded 19488768 of 21187400 bytes (92.0%%, 2.2s
remaining)
[_chunk_report_] Downloaded 20299776 of 21187400 bytes (95.8%%, 1.1s
remaining)
[_chunk_report_] Downloaded 21110784 of 21187400 bytes (99.6%%, 0.1s
remaining)
[fetch_single_file] ...done. (30 seconds, 0 min)
[uncompress_file] Extracting data from
/home/runner/nilearn_data/ibc/75e62c44985852e000c2b2865badf72d/download...
[uncompress_file] .. done.
Define a masker¶
- We define a nilearn masker that will be used to handle relevant data.
For more information, visit : ‘http://nilearn.github.io/manipulating_images/masker_objects.html’
from nilearn.maskers import NiftiMasker
masker = NiftiMasker(mask_img=mask_img).fit()
Prepare the data¶
For each subject, we will use two series of contrasts acquired during two independent sessions with a different phase encoding: Antero-posterior(AP) or Postero-anterior(PA).
# To infer a template for subjects sub-01 to sub-06 for both AP and PA data,
# we make a list of 4D niimgs from our list of list of files containing 3D images
from nilearn.image import concat_imgs
template_train = []
for i in range(5):
template_train.append(concat_imgs(imgs[i]))
# sub-07 (that is 5th in the list) will be our left-out subject.
# We make a single 4D Niimg from our list of 3D filenames.
left_out_subject = concat_imgs(imgs[5])
Compute a baseline (average of subjects)¶
We create an image with as many contrasts as any subject representing for each contrast the average of all train subjects maps.
import numpy as np
masked_imgs = [masker.transform(img) for img in template_train]
average_img = np.mean(masked_imgs, axis=0)
average_subject = masker.inverse_transform(average_img)
Create a template from the training subjects.¶
- We define an estimator using the class TemplateAlignment:
We align the whole brain through ‘multiple’ local alignments.
These alignments are calculated on a parcellation of the brain in 50 pieces, this parcellation creates group of functionnally similar voxels.
The template is created iteratively, aligning all subjects data into a common space, from which the template is inferred and aligning again to this new template space.
from fmralign.template_alignment import TemplateAlignment
# We use Procrustes/scaled orthogonal alignment method
template_estim = TemplateAlignment(
n_pieces=50,
alignment_method="scaled_orthogonal",
masker=masker,
)
template_estim.fit(template_train)
procrustes_template = template_estim.template
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: UserWarning: Overriding provided-default estimator parameters with provided masker parameters :
Parameter mask_strategy :
Masker parameter background - overriding estimator parameter epi
Parameter smoothing_fwhm :
Masker parameter None - overriding estimator parameter 4.0
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: FutureWarning: The nifti_maps_masker_ attribute is deprecated andwill be removed in Nilearn 0.11.3. Please use maps_masker_ instead.
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:189: UserWarning:
Some parcels are more than 1000 voxels wide it can slow down alignment,especially optimal_transport :
parcel 4 : 1988 voxels
parcel 11 : 1402 voxels
parcel 13 : 1028 voxels
parcel 14 : 1353 voxels
parcel 16 : 1890 voxels
parcel 17 : 2191 voxels
parcel 21 : 2491 voxels
parcel 22 : 1588 voxels
parcel 28 : 2337 voxels
parcel 31 : 1809 voxels
parcel 34 : 1904 voxels
parcel 37 : 1485 voxels
parcel 39 : 1741 voxels
parcel 46 : 2855 voxels
parcel 49 : 2602 voxels
parcel 50 : 2311 voxels
warnings.warn(warning)
Predict new data for left-out subject¶
We predict the contrasts of the left-out subject using the template we just created. We use the transform method of the estimator. This method takes the left-out subject as input, computes a pairwise alignment with the template and returns the aligned data.
predictions_from_template = template_estim.transform(left_out_subject)
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/nilearn/masking.py:979: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
return new_img_like(mask_img, unmasked, affine)
Score the baseline and the prediction¶
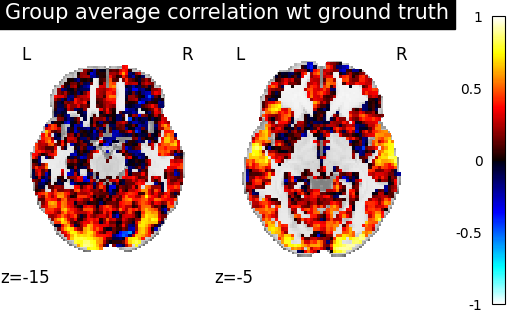
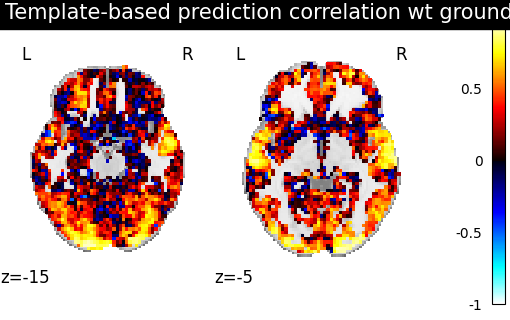
We use a utility scoring function to measure the voxelwise correlation between the images. That is, for each voxel, we measure the correlation between its profile of activation without and with alignment, to see if template-based alignment was able to improve inter-subject similarity.
from fmralign.metrics import score_voxelwise
average_score = masker.inverse_transform(
score_voxelwise(left_out_subject, average_subject, masker, loss="corr")
)
template_score = masker.inverse_transform(
score_voxelwise(
predictions_from_template, procrustes_template, masker, loss="corr"
)
)
Plotting the measures¶
Finally we plot both scores
from nilearn import plotting
baseline_display = plotting.plot_stat_map(
average_score, display_mode="z", vmax=1, cut_coords=[-15, -5]
)
baseline_display.title("Left-out subject correlation with group average")
display = plotting.plot_stat_map(
template_score, display_mode="z", cut_coords=[-15, -5], vmax=1
)
display.title("Aligned subject correlation with Procrustes template")
We observe that creating a template and aligning a new subject to it yields better inter-subject similarity than regular euclidean averaging.
Total running time of the script: (5 minutes 15.011 seconds)