Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Co-smoothing prediction using the Individual Neural Tuning Model¶
In this tutorial, we show how to better predict new contrasts for a target subject using many source subjects corresponding contrasts. For this purpose, we create a template to which we align the target subject, using shared information. We then predict new images for the target and compare them to a baseline.
We mostly rely on Python common packages and on nilearn to handle functional data in a clean fashion.
To run this example, you must launch IPython via ipython
--matplotlib in a terminal, or use jupyter-notebook.
import warnings
warnings.filterwarnings("ignore")
Retrieve the data¶
In this example we use the IBC dataset, which includes a large number of different contrasts maps for 12 subjects. We download the images for subjects sub-01, sub-02, sub-04, sub-05, sub-06 and sub-07 (or retrieve them if they were already downloaded). imgs is the list of paths to available statistical images for each subjects. df is a dataframe with metadata about each of them. mask is a binary image used to extract grey matter regions.
[get_dataset_dir] Dataset found in /home/runner/nilearn_data/ibc
Define a masker¶
- We define a nilearn masker that will be used to handle relevant data.
For more information, visit : ‘https://nilearn.github.io/stable/manipulating_images/masker_objects.html’
from nilearn.maskers import NiftiMasker
masker = NiftiMasker(mask_img=mask_img).fit()
Prepare the data¶
For each subject, we will use two series of contrasts acquired during two independent sessions with a different phase encoding: Antero-posterior(AP) or Postero-anterior(PA).
# To infer a template for subjects sub-01 to sub-06 for both AP and PA data,
# we make a list of 4D niimgs from our list of list of files containing 3D images
from nilearn.image import concat_imgs
template_train = []
for i in range(6):
template_train.append(concat_imgs(imgs[i]))
# For subject sub-07, we split it in two folds:
# - target train: sub-07 AP contrasts, used to learn alignment to template
# - target test: sub-07 PA contrasts, used as a ground truth to score predictions
# We make a single 4D Niimg from our list of 3D filenames
target_train = df[df.subject == "sub-07"][df.acquisition == "ap"].path.values
target_train = concat_imgs(target_train)
target_train_data = masker.transform(target_train)
target_test = df[df.subject == "sub-07"][df.acquisition == "pa"].path.values
Compute a baseline (average of subjects)¶
We create an image with as many contrasts as any subject representing for each contrast the average of all train subjects maps.
import numpy as np
masked_imgs = [masker.transform(img) for img in template_train]
average_img = np.mean(masked_imgs[:-1], axis=0)
average_subject = masker.inverse_transform(average_img)
Create a template from the training subjects.¶
- We define an estimator using the class TemplateAlignment:
We align the whole brain through multiple local alignments.
These alignments are calculated on a parcellation of the brain in 100 pieces, this parcellation creates group of functionnally similar voxels.
The template is created iteratively, aligning all subjects data into a common space, from which the template is inferred and aligning again to this new template space.
from nilearn.image import index_img
from fmralign.alignment_methods import IndividualizedNeuralTuning
from fmralign.hyperalignment.piecewise_alignment import PiecewiseAlignment
from fmralign.hyperalignment.regions import compute_parcels
Predict new data for left-out subject¶
We use target_train data to fit the transform, indicating it corresponds to the contrasts indexed by train_index and predict from this learnt alignment contrasts corresponding to template test_index numbers. For each train subject and for the template, the AP contrasts are sorted from 0, to 53, and then the PA contrasts from 53 to 106.
train_index = range(53)
test_index = range(53, 106)
denoising_data = np.array(masked_imgs)[:, train_index, :]
training_data = np.array(masked_imgs)[:-1]
target_test_masked = np.array(masked_imgs)[:, test_index, :]
parcels = compute_parcels(
niimg=template_train[0], mask=masker, n_parcels=100, n_jobs=5
)
denoiser = PiecewiseAlignment(n_jobs=5)
denoised_signal = denoiser.fit_transform(X=denoising_data, regions=parcels)
target_denoised_data = denoised_signal[-1]
model = IndividualizedNeuralTuning(
parcels=parcels,
)
model.fit(training_data, verbose=False)
stimulus_ = np.copy(model.shared_response)
# From the denoised data and the stimulus, we can now extract the tuning
# matrix from sub-07 AP contrasts, and use it to predict the PA contrasts.
target_tuning = model._tuning_estimator(
shared_response=stimulus_[train_index], target=target_denoised_data
)
# We input the mapping image target_train in a list, we could have input more
# than one subject for which we'd want to predict : [train_1, train_2 ...]
pred = model._reconstruct_signal(
shared_response=stimulus_[test_index], individual_tuning=target_tuning
)
prediction_from_template = masker.inverse_transform(pred)
# As a baseline prediction, let's just take the average of activations across subjects.
prediction_from_average = index_img(average_subject, test_index)
[Loading/Parcel] : Parcellating...
Minimum length parcel: 5
[RegionAlignment] Shape of input data: (6, 53, 46448)
[ParcelAlignment]Computing global template M ...
[Parallel(n_jobs=5)]: Using backend LokyBackend with 5 concurrent workers.
[Parallel(n_jobs=5)]: Done 40 tasks | elapsed: 1.7s
[Parallel(n_jobs=5)]: Done 100 out of 100 | elapsed: 2.0s finished
[Parallel(n_jobs=1)]: Done 49 tasks | elapsed: 7.5s
[Parallel(n_jobs=1)]: Done 100 out of 100 | elapsed: 15.2s finished
Score the baseline and the prediction¶
We use a utility scoring function to measure the voxelwise correlation between the prediction and the ground truth. That is, for each voxel, we measure the correlation between its profile of activation without and with alignment, to see if alignment was able to predict a signal more alike the ground truth.
from fmralign.metrics import score_voxelwise
# Now we use this scoring function to compare the correlation of predictions
# made from group average and from template with the real PA contrasts of sub-07
average_score = masker.inverse_transform(
score_voxelwise(target_test, prediction_from_average, masker, loss="corr")
)
template_score = masker.inverse_transform(
score_voxelwise(target_test, prediction_from_template, masker, loss="corr")
)
Plotting the measures¶
Finally we plot both scores
from nilearn import plotting
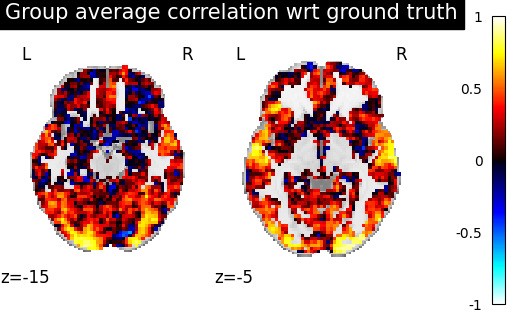
baseline_display = plotting.plot_stat_map(
average_score, display_mode="z", vmax=1, cut_coords=[-15, -5]
)
baseline_display.title("Group average correlation wrt ground truth")
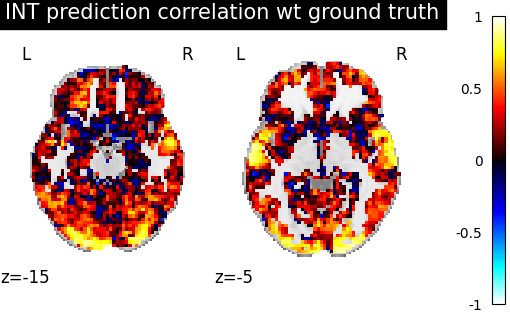
display = plotting.plot_stat_map(
template_score, display_mode="z", cut_coords=[-15, -5], vmax=1
)
display.title("INT prediction correlation wt ground truth")
We observe that creating a template and aligning a new subject to it yields a prediction that is better correlated with the ground truth than just using the average activations of subjects.
plotting.show()
Total running time of the script: (1 minutes 23.568 seconds)