Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Alignment methods benchmark (pairwise ROI case).¶
In this tutorial, we compare various methods of alignment on a pairwise alignment problem for Individual Brain Charting subjects. For each subject, we have a lot of functional informations in the form of several task-based contrast per subject. We will just work here on a ROI.
We mostly rely on python common packages and on nilearn to handle functional data in a clean fashion.
To run this example, you must launch IPython via ipython --matplotlib in
a terminal, or use jupyter-notebook.
Retrieve the data¶
In this example we use the IBC dataset, which include a large number of different contrasts maps for 12 subjects. We download the images for subjects sub-01 and sub-02. Files is the list of paths for each subjects. df is a dataframe with metadata about each of them.
[get_dataset_dir] Dataset found in /home/runner/nilearn_data/ibc
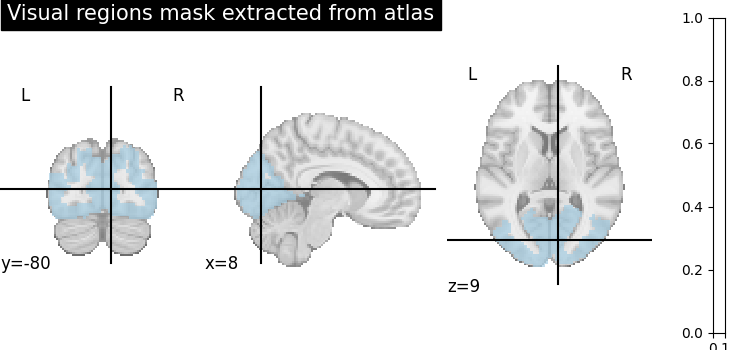
Extract a mask for the visual cortex from Yeo Atlas¶
First, we fetch and plot the complete atlas
from nilearn import datasets, plotting
from nilearn.image import concat_imgs, load_img, new_img_like, resample_to_img
atlas_yeo_2011 = datasets.fetch_atlas_yeo_2011()
atlas = load_img(atlas_yeo_2011.thick_7)
# Select visual cortex, create a mask and resample it to the right resolution
mask_visual = new_img_like(atlas, atlas.get_fdata() == 1)
resampled_mask_visual = resample_to_img(
mask_visual, mask, interpolation="nearest"
)
# Plot the mask we will use
plotting.plot_roi(
resampled_mask_visual,
title="Visual regions mask extracted from atlas",
cut_coords=(8, -80, 9),
colorbar=True,
cmap="Paired",
)
[get_dataset_dir] Dataset found in /home/runner/nilearn_data/yeo_2011
/home/runner/work/fmralign/fmralign/examples/plot_alignment_methods_benchmark.py:47: FutureWarning: 'force_resample' will be set to 'True' by default in Nilearn 0.13.0.
Use 'force_resample=True' to suppress this warning.
resampled_mask_visual = resample_to_img(
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/nilearn/image/resampling.py:805: FutureWarning: From release 0.13.0 onwards, this function will, by default, copy the header of the input image to the output. Currently, the header is reset to the default Nifti1Header. To suppress this warning and use the new behavior, set `copy_header=True`.
return resample_img(
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1782e8f910>
Define a masker¶
We define a nilearn masker that will be used to handle relevant data. For more information, visit : ‘http://nilearn.github.io/manipulating_images/masker_objects.html’
from nilearn.maskers import NiftiMasker
roi_masker = NiftiMasker(mask_img=resampled_mask_visual).fit()
Prepare the data¶
For each subject, for each task and conditions, our dataset contains two independent acquisitions, similar except for one acquisition parameter, the encoding phase used that was either Antero-Posterior (AP) or Postero-Anterior (PA). Although this induces small differences in the final data, we will take advantage of these pseudo-duplicates to create a training and a testing set that contains roughly the same signals but acquired independently.
# The training set, used to learn alignment from source subject toward target:
# * source train: AP contrasts for subject sub-01
# * target train: AP contrasts for subject sub-02
#
source_train = concat_imgs(
df[df.subject == "sub-01"][df.acquisition == "ap"].path.values
)
target_train = concat_imgs(
df[df.subject == "sub-02"][df.acquisition == "ap"].path.values
)
# The testing set:
# * source test: PA contrasts for subject one, used to predict
# the corresponding contrasts of subject sub-01
# * target test: PA contrasts for subject sub-02, used as a ground truth
# to score our predictions
#
source_test = concat_imgs(
df[df.subject == "sub-01"][df.acquisition == "pa"].path.values
)
target_test = concat_imgs(
df[df.subject == "sub-02"][df.acquisition == "pa"].path.values
)
/home/runner/work/fmralign/fmralign/examples/plot_alignment_methods_benchmark.py:88: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
df[df.subject == "sub-01"][df.acquisition == "ap"].path.values
/home/runner/work/fmralign/fmralign/examples/plot_alignment_methods_benchmark.py:91: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
df[df.subject == "sub-02"][df.acquisition == "ap"].path.values
/home/runner/work/fmralign/fmralign/examples/plot_alignment_methods_benchmark.py:102: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
df[df.subject == "sub-01"][df.acquisition == "pa"].path.values
/home/runner/work/fmralign/fmralign/examples/plot_alignment_methods_benchmark.py:105: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
df[df.subject == "sub-02"][df.acquisition == "pa"].path.values
Choose the number of regions for local alignment¶
First, as we will proceed to local alignment we choose a suitable number of regions so that each of them is approximately 200 voxels wide. Then our estimator will first make a functional clustering of voxels based on train data to divide them into meaningful regions.
The chosen region of interest contains 6631.0 voxels
We will cluster them in 33 regions
Define the estimators, fit them and do a prediction¶
- On each region, we search for a transformation R that is either :
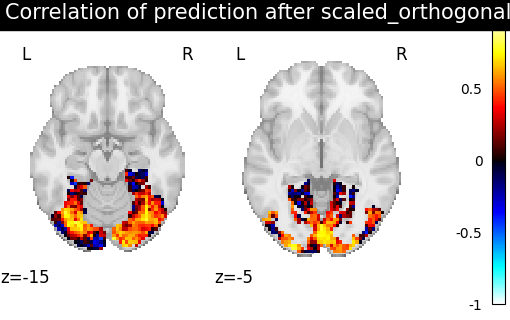
orthogonal, i.e. R orthogonal, scaling sc s.t. ||sc RX - Y ||^2 is minimized
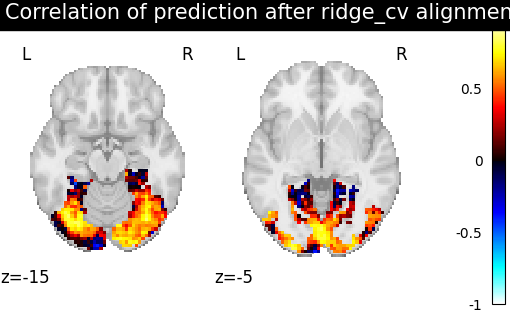
a ridge regression : ||XR - Y||^2 + alpha *||R||^2 with a L2 penalization on the norm of R.
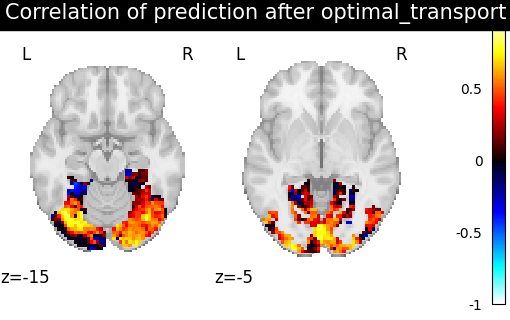
- the optimal transport plan, which yields the minimal transport cost
while respecting the mass conservation constraints. Calculated with entropic regularization.
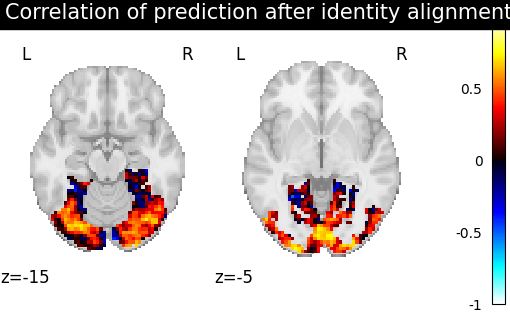
we also include identity (no alignment) as a baseline.
Then for each method we define the estimator fit it, predict the new image and plot its correlation with the real signal.
from fmralign.metrics import score_voxelwise
from fmralign.pairwise_alignment import PairwiseAlignment
methods = ["identity", "scaled_orthogonal", "ridge_cv", "optimal_transport"]
for method in methods:
alignment_estimator = PairwiseAlignment(
alignment_method=method, n_pieces=n_pieces, masker=roi_masker
)
alignment_estimator.fit(source_train, target_train)
target_pred = alignment_estimator.transform(source_test)
# derive correlation between prediction, test
method_error = score_voxelwise(
target_test, target_pred, masker=roi_masker, loss="corr"
)
# plot correlation for each method
aligned_score = roi_masker.inverse_transform(method_error)
title = f"Correlation of prediction after {method} alignment"
display = plotting.plot_stat_map(
aligned_score,
display_mode="z",
cut_coords=[-15, -5],
vmax=1,
title=title,
)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: UserWarning: Overriding provided-default estimator parameters with provided masker parameters :
Parameter mask_strategy :
Masker parameter background - overriding estimator parameter epi
Parameter smoothing_fwhm :
Masker parameter None - overriding estimator parameter 4.0
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: FutureWarning: The nifti_maps_masker_ attribute is deprecated andwill be removed in Nilearn 0.11.3. Please use maps_masker_ instead.
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:189: UserWarning:
Some parcels are more than 1000 voxels wide it can slow down alignment,especially optimal_transport :
parcel 1 : 1725 voxels
parcel 15 : 1484 voxels
parcel 16 : 1190 voxels
parcel 23 : 1048 voxels
warnings.warn(warning)
/home/runner/work/fmralign/fmralign/fmralign/metrics.py:62: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
pearsonr(X_gt[:, vox], X_pred[:, vox])[
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/nilearn/plotting/img_plotting.py:1416: UserWarning: Non-finite values detected. These values will be replaced with zeros.
safe_get_data(stat_map_img, ensure_finite=True),
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: UserWarning: Overriding provided-default estimator parameters with provided masker parameters :
Parameter mask_strategy :
Masker parameter background - overriding estimator parameter epi
Parameter smoothing_fwhm :
Masker parameter None - overriding estimator parameter 4.0
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: FutureWarning: The nifti_maps_masker_ attribute is deprecated andwill be removed in Nilearn 0.11.3. Please use maps_masker_ instead.
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:189: UserWarning:
Some parcels are more than 1000 voxels wide it can slow down alignment,especially optimal_transport :
parcel 1 : 1725 voxels
parcel 15 : 1484 voxels
parcel 16 : 1190 voxels
parcel 23 : 1048 voxels
warnings.warn(warning)
/home/runner/work/fmralign/fmralign/fmralign/metrics.py:62: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
pearsonr(X_gt[:, vox], X_pred[:, vox])[
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/nilearn/plotting/img_plotting.py:1416: UserWarning: Non-finite values detected. These values will be replaced with zeros.
safe_get_data(stat_map_img, ensure_finite=True),
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: UserWarning: Overriding provided-default estimator parameters with provided masker parameters :
Parameter mask_strategy :
Masker parameter background - overriding estimator parameter epi
Parameter smoothing_fwhm :
Masker parameter None - overriding estimator parameter 4.0
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: FutureWarning: The nifti_maps_masker_ attribute is deprecated andwill be removed in Nilearn 0.11.3. Please use maps_masker_ instead.
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:189: UserWarning:
Some parcels are more than 1000 voxels wide it can slow down alignment,especially optimal_transport :
parcel 1 : 1725 voxels
parcel 15 : 1484 voxels
parcel 16 : 1190 voxels
parcel 23 : 1048 voxels
warnings.warn(warning)
/home/runner/work/fmralign/fmralign/fmralign/metrics.py:62: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
pearsonr(X_gt[:, vox], X_pred[:, vox])[
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/nilearn/plotting/img_plotting.py:1416: UserWarning: Non-finite values detected. These values will be replaced with zeros.
safe_get_data(stat_map_img, ensure_finite=True),
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: UserWarning: Overriding provided-default estimator parameters with provided masker parameters :
Parameter mask_strategy :
Masker parameter background - overriding estimator parameter epi
Parameter smoothing_fwhm :
Masker parameter None - overriding estimator parameter 4.0
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:258: FutureWarning: The nifti_maps_masker_ attribute is deprecated andwill be removed in Nilearn 0.11.3. Please use maps_masker_ instead.
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/fmralign/_utils.py:189: UserWarning:
Some parcels are more than 1000 voxels wide it can slow down alignment,especially optimal_transport :
parcel 1 : 1725 voxels
parcel 15 : 1484 voxels
parcel 16 : 1190 voxels
parcel 23 : 1048 voxels
warnings.warn(warning)
/home/runner/work/fmralign/fmralign/fmralign/metrics.py:62: ConstantInputWarning: An input array is constant; the correlation coefficient is not defined.
pearsonr(X_gt[:, vox], X_pred[:, vox])[
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/nilearn/plotting/img_plotting.py:1416: UserWarning: Non-finite values detected. These values will be replaced with zeros.
safe_get_data(stat_map_img, ensure_finite=True),
We can observe that all alignment methods perform better than identity (no alignment). Ridge is the best performing method, followed by Optimal Transport. If you use Ridge though, be careful about the smooth predictions it yields.
Total running time of the script: (0 minutes 48.277 seconds)